Distribution-based clustering algorithms, also known as probabilistic clustering algorithms, are a class of machine learning algorithms that assume that the data points are generated from a mixture of probability distributions. These algorithms aim to identify the underlying probability distributions that generate the data, and use this information to cluster the data into groups with similar properties.
One common distribution-based clustering algorithm is the Gaussian Mixture Model (GMM). GMM assumes that the data points are generated from a mixture of Gaussian distributions, and aims to estimate the parameters of these distributions, including the means and covariances of each distribution. Let’s see below what is GMM in ML and how we can implement in Python programming language.
Gaussian Mixture Model
Gaussian Mixture Models (GMM) is a popular clustering algorithm used in machine learning that assumes that the data is generated from a mixture of Gaussian distributions. In other words, GMM tries to fit a set of Gaussian distributions to the data, where each Gaussian distribution represents a cluster in the data.
GMM has several advantages over other clustering algorithms, such as the ability to handle overlapping clusters, model the covariance structure of the data, and provide probabilistic cluster assignments for each data point. This makes GMM a popular choice in many applications, such as image segmentation, pattern recognition, and anomaly detection.

Implementation in Python
In Python, the Scikit-learn library provides the GaussianMixture class for implementing the GMM algorithm. The class takes several parameters, including the number of components (i.e., the number of clusters to identify), the covariance type, and the initialization method.
Here is an example of how to implement GMM using the Scikit-learn library in Python −
Example
from sklearn.mixture import GaussianMixture
from sklearn.datasets import make_blobs
import matplotlib.pyplot as plt
# generate a dataset
X, _ = make_blobs(n_samples=200, centers=4, random_state=0)# create an instance of the GaussianMixture class
gmm = GaussianMixture(n_components=4)# fit the model to the dataset
gmm.fit(X)# predict the cluster labels for the data points
labels = gmm.predict(X)# print the cluster labelsprint("Cluster labels:", labels)
plt.figure(figsize=(7.5,3.5))
plt.scatter(X[:,0], X[:,1], c=labels, cmap='viridis')
plt.show()In this example, we first generate a synthetic dataset using the make_blobs() function from Scikit-learn. We then create an instance of the GaussianMixture class with 4 components and fit the model to the dataset using the fit() method. Finally, we predict the cluster labels for the data points using the predict() method and print the resulting labels.
Output
When you execute this program, it will produce the following plot as the output −
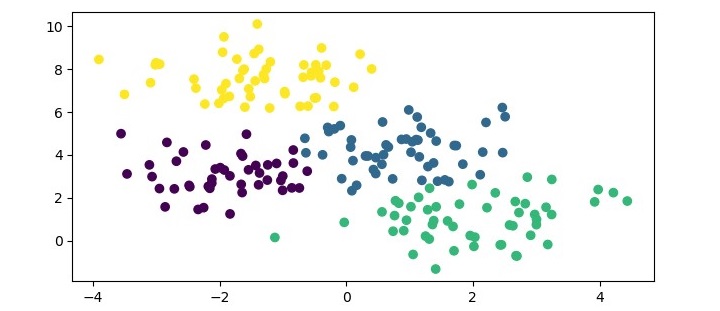
In addition, you will get the following output on the terminal −
Cluster labels: [2 0 1 3 2 1 0 1 1 1 1 2 0 0 2 1 3 3 3 1 3 1 2 0 2 2 3 2 2 1 3 1 0 2 0 1 0
1 1 3 3 3 3 1 2 0 1 3 3 1 3 0 0 3 2 3 0 2 3 2 3 1 2 1 3 1 2 3 0 0 2 2 1 1
0 3 0 0 2 2 3 1 2 2 0 1 1 2 0 0 3 3 3 1 1 2 0 3 2 1 3 2 2 3 3 0 1 2 2 1 3
0 0 2 2 1 2 0 3 1 3 0 1 2 1 0 1 0 2 1 0 2 1 3 3 0 3 3 2 3 2 0 2 2 2 2 1 2
0 3 3 3 1 0 2 1 3 0 3 2 3 2 2 0 0 3 1 2 2 0 1 1 0 3 3 3 1 3 0 0 1 2 1 2 1
0 0 3 1 3 2 2 1 3 0 0 0 1 3 1]
The covariance type parameter in GMM controls the type of covariance matrix to use for the Gaussian distributions. The available options include “full” (full covariance matrix), “tied” (tied covariance matrix for all clusters), “diag” (diagonal covariance matrix), and “spherical” (a single variance parameter for all dimensions). The initialization method parameter controls the method used to initialize the parameters of the Gaussian distributions.
Explore our latest online courses and learn new skills at your own pace. Enroll and become a certified expert to boost your career.
Advantages of Gaussian Mixture Models
Following are the advantages of using Gaussian Mixture Models −
- Gaussian Mixture Models (GMM) can model arbitrary distributions of data, making it a flexible clustering algorithm.
- It can handle datasets with missing or incomplete data.
- It provides a probabilistic framework for clustering, which can provide more information about the uncertainty of the clustering results.
- It can be used for density estimation and generation of new data points that follow the same distribution as the original data.
- It can be used for semi-supervised learning, where some data points have known labels and are used to train the model.

Disadvantages of Gaussian Mixture Models
Following are some of the disadvantages of using Gaussian Mixture Models −
- GMM can be sensitive to the choice of initial parameters, such as the number of clusters and the initial values for the means and covariances of the clusters.
- It can be computationally expensive for high-dimensional datasets, as it involves computing the inverse of the covariance matrix, which can be expensive for large matrices.
- It assumes that the data is generated from a mixture of Gaussian distributions, which may not be true for all datasets.
- It may be prone to overfitting, especially when the number of parameters is large or the dataset is small.
- It can be difficult to interpret the resulting clusters, especially when the covariance matrices are complex.
Leave a Reply